Lecture 09: Bayesian Regression
Classical Linear regression
\[ \newcommand{\mc}[1]{\mathcal{#1}} \newcommand{\R}{\mathbb{R}} \newcommand{\E}{\mathbb{E}} \renewcommand{\P}{\mathbb{P}} \newcommand{\var}{{\rm Var}} % Variance \newcommand{\mse}{{\rm MSE}} % MSE \newcommand{\bias}{{\rm Bias}} % MSE \newcommand{\cov}{{\rm Cov}} % Covariance \newcommand{\iid}{\stackrel{\rm iid}{\sim}} \newcommand{\ind}{\stackrel{\rm ind}{\sim}} \renewcommand{\choose}[2]{\binom{#1}{#2}} % Choose \newcommand{\chooses}[2]{{}_{#1}C_{#2}} % Small choose \newcommand{\cd}{\stackrel{d}{\rightarrow}} \newcommand{\cas}{\stackrel{a.s.}{\rightarrow}} \newcommand{\cp}{\stackrel{p}{\rightarrow}} \newcommand{\bin}{{\rm Bin}} \newcommand{\ber}{{\rm Ber}} \DeclareMathOperator*{\argmax}{argmax} \DeclareMathOperator*{\argmin}{argmin} \]
Linear regression
- Suppose we have samples \(\{x_i, y_i\}_{i=1}^n\) where \(y_i \in \R\) and \(x_i \in \R^{p-1}\). We want to model the relationship between \(x_i\) and \(y_i\).
- The classical linear regression model is \(Y = X^T\beta\).
- We can fit the model by the method of ordinary least-square (OLS), i.e., minimizing \(\sum_{i=1}^n (y_i - x_i^T\beta)^2\).
- The solution is \(\hat{\beta} = (X^TX)^{-1}X^TY\), where \(Y = [y_1,\ldots, y_n]^T\) and \[ X = \left[\begin{array}{cc} 1 & x_1^T \\ \vdots & \vdots \\ 1 & x_n^T \end{array} \right]_{n\times p}. \]
- If we further assume \(Y = X^T\beta + \varepsilon\), \(\varepsilon \sim N(0, \sigma^2)\), then the OLS solution \(\hat{\beta} = (X^TX)^{-1}X^TY\) is the MLE of \(\beta\).
Normal linear model
- The simple linear regression model is a normal linear model \[ Y \mid X, \beta, \sigma^2 \sim N(X\beta, \sigma^2I_n) \] where \(I_n\) is the \(n \times n\) identity matrix, \(X \in \R^{n \times p}\), and \(\beta \in \R^p\).
- Both \(\beta\) and \(\sigma^2\) are unknown parameters.
- In practice, it is very likely that the normality assumption is violated. In such cases, we can apply some transformations to \(Y\), e.g., Box-Cox transformation, to make it normal.
- In the next lecture, we will discuss generalized linear models (GLM), i.e., \[ Y \mid X, \beta \sim F_{\eta} \] where \(F_\eta\) is an exponential family and \(\eta = X^T\beta\).
Standard noninformative prior
- A convenient noninformative prior is the uniform prior on \((\beta, \log \sigma)\) or, equivalently, \[ \pi\left(\beta, \sigma^2 \mid X\right) \propto \sigma^{-2} \]
- This prior is useful when there are many samples and only a few parameters, i.e., \(n \gg p\).
Posterior distributions
- Similar to the normal model with unknown mean and variance, the joint posterior can be factorized: \(\pi(\beta, \sigma^2 \mid X, Y) = \pi(\beta \mid \sigma^2, X, Y)\pi(\sigma^2 \mid X, Y)\)
- Conditional posterior distribution of \(\beta\), given \(\sigma\): \[ \beta \mid \sigma^2, y \sim N\left(\hat{\beta}, V_\beta \sigma^2\right) \] where \(\hat{\beta} = (X^TX)^{-1}X^TY\) and \(V_\beta = (X^TX)^{-1}\).
- Marginal posterior distribution of \(\sigma^2\): \[ \sigma^2 \mid y \sim \text{Inv}-\chi^2\left(n-p, s^2\right) \] where \(s^2 = \frac{1}{n-p}(Y - X\hat{\beta})^T(Y - X\hat{\beta})\).
- \(\text{Inv}-\chi^2(\nu, s)\) is the scaled inverse \(\chi^2\) distribution.
- Hence the posterior means of \(\beta\) and \(\sigma^2\) are the same as the OLS estimates.
Posterior predictive distribution
- Suppose we observe a new sample \(X^*\) and we want to predict its corresponding response \(Y^*\).
- The posterior predictive distribution is \[ p(Y^* \mid X^*, X, Y) = \int p(Y^* \mid \beta, \sigma^2, X^*) \pi(\beta, \sigma^2 \mid X, Y) \; d\beta\; d\sigma^2. \]
- For a normal linear model with the noninformative prior \(\pi(\beta, \sigma^2) \propto \sigma^{-2}\), we can compute the analytic form of the predictive distribution.
- However, there is no need to do so, since once we have the posterior samples of \(\beta\) and \(\sigma^2\), we can easily draw samples from the posterior predictive distribution.
- That is, with \((\beta^{(i)}, \sigma^{2,{(i)}}) \iid \pi(\beta, \sigma^2 \mid X, Y)\), we can draw \[ Y^{*(i)} \mid X^*, X, Y \sim N(X^{*T}\beta^{(i)}, \sigma^{2,(i)}). \]
Computation
There are many R packages for fitting Bayesian regression models:
rstanarm: pre-compiled Stan regression models; easier to use; faster; less flexiblebrms: compile models on the fly; slower; more flexiblebayesplot: visualization of Bayesian models- MCMC diagnostics (using functions
mcmc_*) - Posterior prediction checking (PPC) (using functions
ppc_*) - Posterior prediction distribution (PPD) (using functions
ppd_*)
- MCMC diagnostics (using functions
loo: efficient approximate leave-one-out cross-validation for fitted Bayesian modelsshinystan: Interactive diagnostic tools for assessing Bayesian models
Example: Kids’ Test Scores1
Data from a survey of adult American women and their children (a subsample from the National Longitudinal Survey of Youth).
- Source: Gelman and Hill (2007)
- 434 obs. of 4 variables
kid_score: Child’s IQ scoremom_hs: Indicator for whether the mother has a high school degreemom_iq: Mother’s IQ scoremom_age: Mother’s age
Fit a Bayesian linear regression model
Summary of the fitted model
stan_glm
family: gaussian [identity]
formula: kid_score ~ mom_hs + mom_iq
observations: 434
predictors: 3
------
Median MAD_SD
(Intercept) 25.8 5.5
mom_hs 6.0 2.2
mom_iq 0.6 0.1
Auxiliary parameter(s):
Median MAD_SD
sigma 18.2 0.6
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanregPrior Summary
MCMC Diagnostics

MCMC Diagnostic

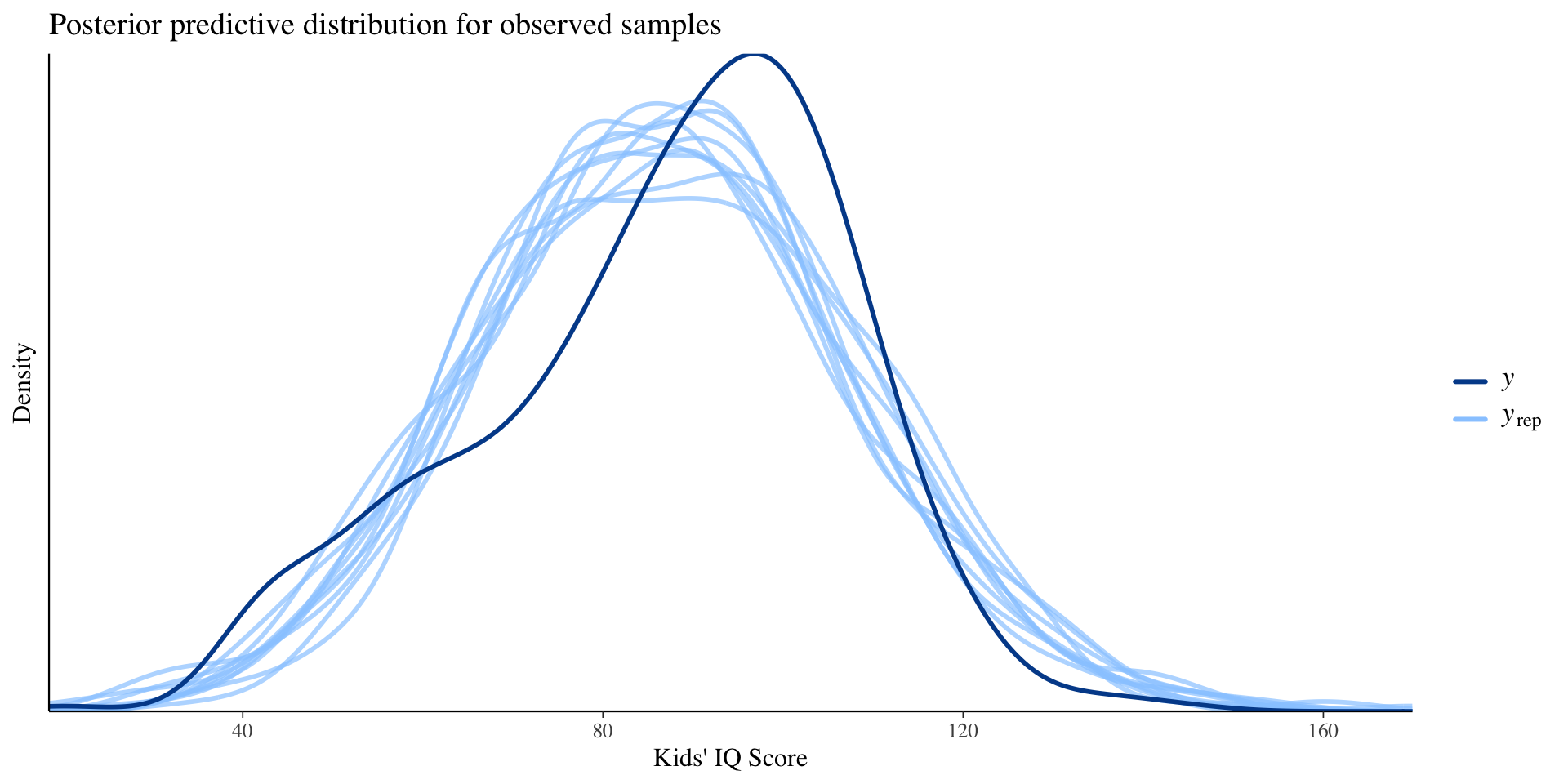
Posterior Prediction Checks (In-sample checking)
Compare observed data to draws from the posterior predictive distribution.
Posterior Prediction Checks (In-sample checking)
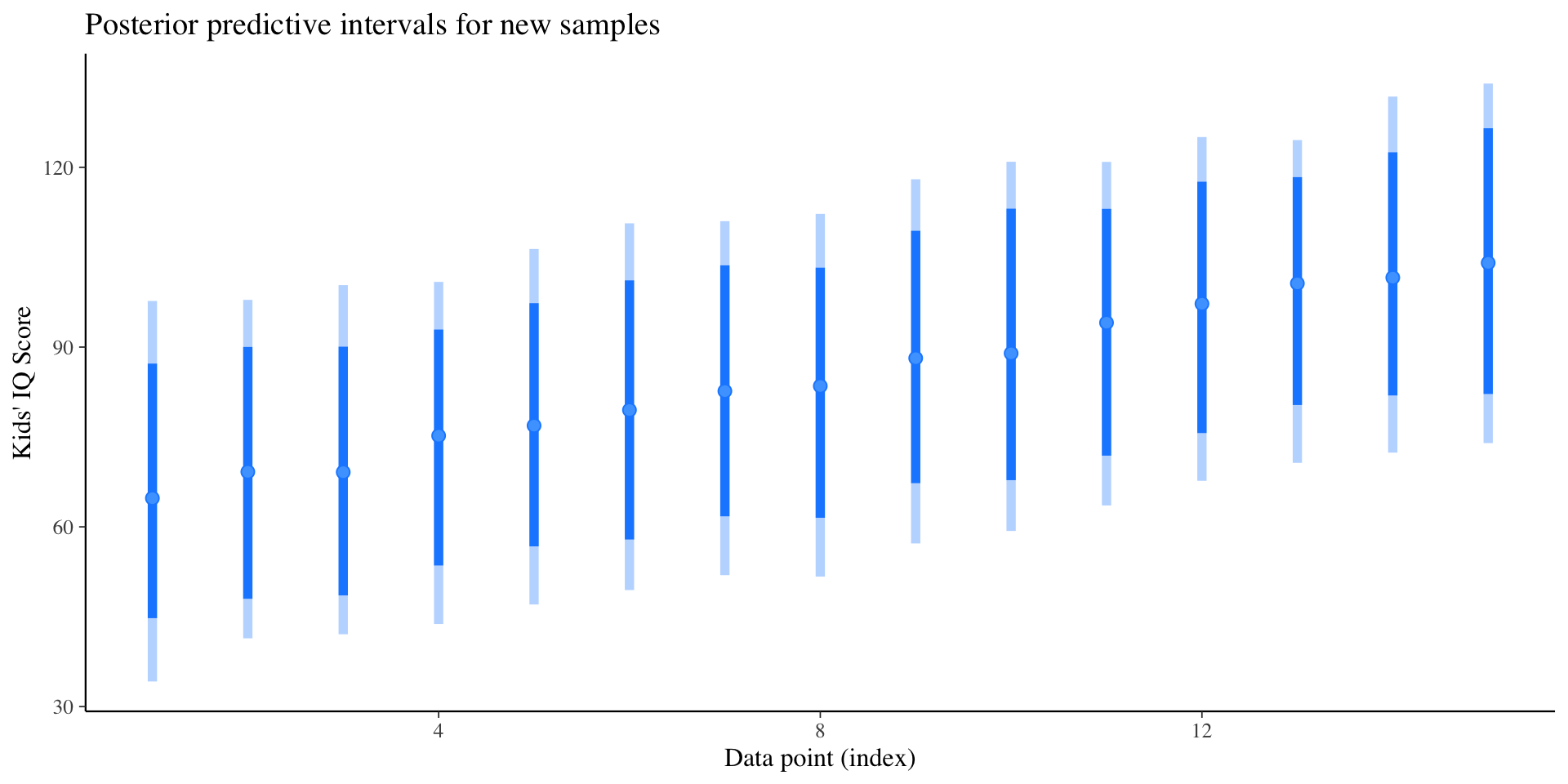
Posterior Prediction Distributions (Out-of-sample prediction)
Compute the posterior predictive distribution for a new sample.
Posterior Prediction Distributions (Out-of-sample prediction)
Interactive tool: shinystan

Regularized regression
Regularized linear regression
- The OLS solution \(\hat{\beta} = (X^TX)^{-1}X^TY\) is obtained by minimizing the residual sum of squares (RSS), i.e., \(\arg\min_{\beta\in\R^p} \|Y - X\beta\|^2\).
- In some cases, the matrix \(X^TX\) is ill-conditioned, i.e., the inverse \((X^TX)^{-1}\) is numerically unstable.
- Also when \(p > n\), the matrix \(X^TX\) is singular and the OLS solution does not exist.
- We can consider the following penalized least-square problem: \[ \arg\min_{\beta\in\R^p} \|Y - X\beta\|^2 + \lambda \cdot\text{Penalty}(\beta) \quad \text{for some } \lambda > 0. \]
- There are three commonly used penalties:
- Ridge regression: \(\text{Penalty}(\beta) = \|\beta\|^2 = \sum_{j=1}^p \beta_j^2\)
- LASSO: \(\text{Penalty}(\beta) = \|\beta\|_1 = \sum_{j=1}^p |\beta_j|\)
- Elastic net: \(\text{Penalty}(\beta) = \alpha \|\beta\|^2 + (1-\alpha) \|\beta\|_1\) for some \(\alpha \in [0, 1]\).
Bayesian interpretation of regularization
- Any penalized likelihood estimator has a Bayesian interpretation, i.e., it is the posterior mode under the prior \(\pi(\beta) \propto \exp(-\lambda\cdot\text{Penalty}(\beta))\).
- For example,
- Ridge regression: normal prior \(\beta_j \sim N(0, \lambda^{-1})\)
- LASSO: Laplace prior \(\beta_j \sim \text{Laplace}(0, \lambda^{-1})\)
- Elastic net: \(\pi(\boldsymbol{\beta}) \propto \exp \left\{-\lambda_1\|\boldsymbol{\beta}\|_1-\lambda_2\|\boldsymbol{\beta}\|_2^2\right\}\)
- Usually, the tuning parameter \(\lambda\) is chosen by cross-validation.
- Under the Bayesian framework, we can
- put a prior on \(\lambda\) (fully Bayesian)
- put a hierarchical prior on \(\lambda\) (hierarchical Bayes)
- estimate it from the data (empirical Bayes)
Bayesian LASSO
LASSO = “Least Absolute Shrinkage and Selection Operator”
Model (this is how
stan_glmimplements LASSO prior): \[\begin{align*} Y \mid X, \beta, \sigma^2 & \sim N(X\beta, \sigma^2 I_n) \\ \beta_j & \iid \text{Laplace}(0, \lambda^{-1}) \\ \lambda & \sim \chi^2_{\nu} \end{align*}\] where \(\nu\) is the degrees of freedom (the default is \(\nu = 1\)).Bias increases as \(\lambda\) increases (producing more zeroes).
Variance decreases as \(\lambda\) increases.
Hence larger value of \(\nu\) produces more shrinkage.
Bayesian LASSO
data <- read.csv("dataset/car_price/CarPrice_Assignment.csv")
fit_lasso <- stan_glm(price ~ highwaympg + citympg + horsepower +
peakrpm + compressionratio,
data = data,
prior = lasso(df = 1, location = 0,
scale = 0.5, autoscale = TRUE),
prior_intercept = normal(location = 0, scale = 1,
autoscale = TRUE),
prior_aux = exponential(rate = 1, autoscale = TRUE),
chains = 4, iter = 2000, seed = 2023,
refresh = 0)Summary of the fitted model
Model Info:
function: stan_glm
family: gaussian [identity]
formula: price ~ highwaympg + citympg + horsepower + peakrpm + compressionratio
algorithm: sampling
sample: 4000 (posterior sample size)
priors: see help('prior_summary')
observations: 205
predictors: 6
Estimates:
mean sd 10% 50% 90%
(Intercept) 9493.2 4656.5 3681.0 9573.9 15322.0
highwaympg -328.0 153.9 -528.2 -320.4 -140.2
citympg 71.7 171.2 -131.5 57.3 296.4
horsepower 139.0 12.4 122.9 139.2 154.7
peakrpm -1.4 0.7 -2.3 -1.4 -0.5
compressionratio 451.8 88.6 338.3 450.3 564.1
sigma 4123.4 212.5 3858.6 4113.1 4398.4
Fit Diagnostics:
mean sd 10% 50% 90%
mean_PPD 13262.4 398.9 12755.5 13256.9 13779.3
The mean_ppd is the sample average posterior predictive distribution of the outcome variable (for details see help('summary.stanreg')).
MCMC diagnostics
mcse Rhat n_eff
(Intercept) 72.0 1.0 4179
highwaympg 2.6 1.0 3374
citympg 2.9 1.0 3555
horsepower 0.2 1.0 4133
peakrpm 0.0 1.0 4251
compressionratio 1.4 1.0 4222
sigma 3.2 1.0 4291
mean_PPD 6.4 1.0 3869
log-posterior 0.1 1.0 801
For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1).High-dimensional linear regression
High-dimensional linear regression
- In some applications, we might have more predictors than samples, i.e., \(p \gg n\).
- In such cases, the sparsity assumption is often used, i.e., only a few predictors are relevant.
- LASSO or Bayesian Lasso can be used to select the relevant predictors.
- However LASSO or Bayesian LASSO have some problems:
- it does not handle correlated variables well
- it shrinks all coefficients with the same intensity
- it often overshrinks large coefficients.
- We will now introduce two priors for high-dimensional sparse linear regression that address these problems.
Spike-and-slab prior
- The spike-and-slab prior1 is \[ \beta_j \mid w, \tau^2 \sim (1 - w)\delta_0 + w \cdot N(0, \tau^2). \]
- Equivalently, \[\begin{align*} \beta_j \mid \gamma_j = 0 & \sim \delta_0 \\ \beta_j \mid \gamma_j = 1 & \sim N(0, \tau^2)\\ \gamma_j & \iid \text{Ber}(w) \end{align*}\]
- \(w\) is the parameter that controls the sparsity, and \(\gamma_j\) is the indicator of the \(j\)-th predictor being relevant.
- This prior is considered the ``gold standard’’ for sparse regression.
- However, it is computationally expensive to update the discrete variables \(\gamma_j\).
Horseshoe prior
- The horseshoe prior1 is \[\begin{align*} \beta_j \mid \lambda_j, \tau & \sim N\left(0, \tau^2 \lambda_j^2\right) \\ \lambda_j & \sim C^{+}(0,1), \quad j=1, \ldots, p \end{align*}\] where \(C^{+}(0, 1)\) is the half-Cauchy distribution.
- This is an example of global-local shrinkage prior, where \(\lambda_j\) is the local shrinkage parameter and \(\tau\) is the global shrinkage parameter.
Available priors for stan_glm
- Student-\(t\) family: cauchy, \(t\), normal
- Hierarchical shrinkage family: horseshoe, horseshoe+
- Laplace family: laplace, lasso
- R2 prior: This prior hinges on prior beliefs about the location of \(R^2\), the proportion of variance in the outcome attributable to the predictors.
- Check https://mc-stan.org/rstanarm/reference/priors.html for more details.
Compare different models
- Suppose we have two regression models: \[\begin{align*} & Y = \beta_0 + \beta_1X_1\\ & Y = \beta_0 + \beta_1X_1 + \beta_2X_2. \end{align*}\]
- We can compare these two models using the Bayes factor.
- However, since we are fitting linear models, we can also use the \(R^2\) to compare the models.
- The \(R^2\) statistic is defined as \[\begin{align*} R^2 & = \frac{\text{Explained variance}}{\text{Total variance}} = 1 - \frac{\text{Residual variance}}{\text{Total variance}}\\ & = 1 - \frac{\sum_{i=1}^n (y_i - \hat{y}_i)^2}{\sum_{i=1}^n (y_i - \bar{y})^2}. \end{align*}\]
- Therefore, a model with a larger \(R^2\) is preferred.
Bayesian \(R^2\)
- However, the \(R^2\) always increases even if we add irrelevant predictors.
- Adjusted \(R^2\) is a modification of \(R^2\) that penalizes the addition of irrelevant predictors \[ R^2_{\text{adj}} =1-\left(1-R^2\right) \frac{n-1}{n-p-1}. \]
- Instead of comparing the \(R^2\) or the adjusted \(R^2\) of the two models, we can compare the posterior distributions of the \(R^2\) of the two models.
Example
fit1 <- stan_glm(kid_score ~ mom_hs, data=kidiq,
seed=2023, refresh=0)
fit2 <- stan_glm(kid_score ~ mom_hs + mom_iq, data=kidiq,
seed=2023, refresh=0)
## Adding random noise as predictors
set.seed(2023)
n <- nrow(kidiq)
kidiqr <- kidiq
kidiqr$noise <- array(rnorm(5*n), c(n,5))
fit3 <- stan_glm(kid_score ~ mom_hs + mom_iq + noise, data=kidiqr,
seed=2023, refresh=0)
r2_fit1 <- bayes_R2(fit1)
r2_fit2 <- bayes_R2(fit2)
r2_fit3 <- bayes_R2(fit3)Posterior distribution of \(R^2\)
Although adding noise predictors does increase the \(R^2\), the change is negligible compared to the uncertainty of \(R^2\).
